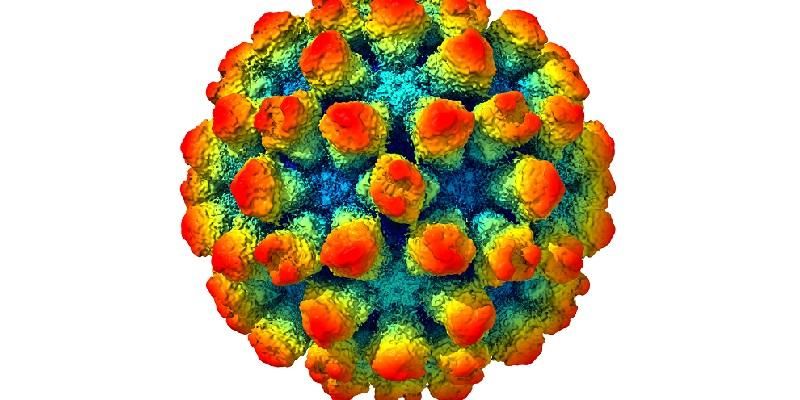
Every picture tells a story… none more so than this detailed visualisation of a strain of the norovirus.
Created from 13,000 separate images taken by an electron microscope, it reveals in rich detail the structure of the virus. It shows bump-like protrusions on the outside of the virus capsid, the protein shell that holds the genome.
Joseph Snowden, a PhD researcher from the Astbury Centre for Structural Molecular Biology , said it revealed that the protein casing did not keep a fixed shape, as the protrusions on the surface would extend, retract and rotate.
This dynamic shape shifting may hold clues as to why the noroviruses are such potent pathogens, responsible for over 200,000 deaths worldwide each year, mainly in low-to-middle-income countries. In the UK, noroviruses cause the winter vomiting bug that forces the closure of schools and hospitals.
To identify subtle changes to the structure of these protein shells, the scientists processed the data from the images with a super computer, using a method called “focussed classification”.
The study, Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM, is reported in the online journal PLOS Biology.
For safety reasons, the researchers used a mouse norovirus in their study, which closely resembles the human noroviruses.
Mr Snowden said: “The constant changing or morphing of the virus shell may enable it to confuse the body’s defence systems.
“Immune systems work on the basis of molecular shape. The body’s defences will produce proteins that bind to pathogens to prevent them from infecting host cells. But if the shape of the virus is constantly changing, the body’s defence systems may be unable to act efficiently.”
The protrusions seem able to move independently or in a co-ordinated fashion and the scientists believe this may enable the virus to prime itself ready for infection depending on where it is, for example if it is in the digestive tract of a host organism.
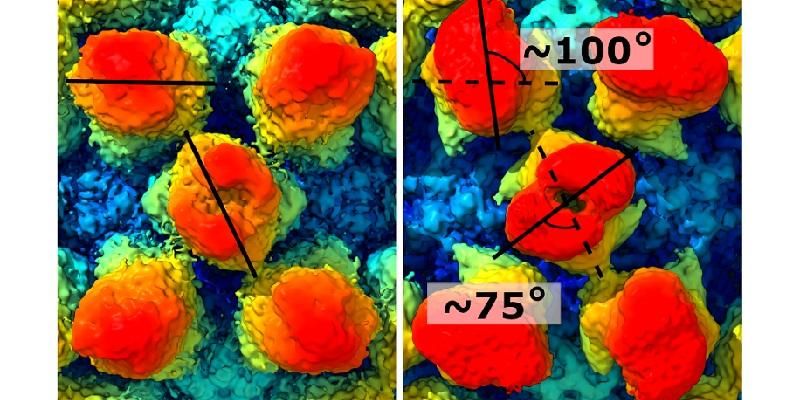
Two images side by side reveal how the bump-like protrusions have rotated. The one on the top left hand side by about 100 degrees. The one in the centre by about 27 degrees.
It is believed that this study is the first study to use the focussed classification computing tool to investigate the structure of a norovirus.
Structural insight to aid vaccine development
Dr Morgan Herod, from the School of Molecular and Cellular Biology at Leeds and one of the senior authors on this study, hopes that a greater understanding of the structure of the norovirus may help with vaccine development, which so far has been unsuccessful.
Dr Herod said “The University of Leeds has a strong track record in the use of virus-like particles (VLPs) to create vaccine candidates, for example against polio. VLPs are harmless proteins modelled on the structure of a virus’s casing and they fool the immune system into thinking the body is under attack, prompting an immune response. “
“Our research shows that the capsid or protein shell of norovirus is dynamic – and perhaps we have to look at VLPs that are better able to mimic this aspect of norovirus structure.”
“That will make the vaccine development task a little harder.”
Further information
The research was funded by Wellcome, the Medical Research Council and the Biotechnology and Biological Sciences Research Council. The electron microscopy was performed in the Astbury Biostructure Laboratory, which was funded by the University of Leeds and Wellcome. The funders had no role in study design, data collection and analysis, decisions to publish, or preparation of the manuscript.
For further information, please contact David Lewis in the University of Leeds press office: d.lewis@leeds.ac.uk