The complex and mysterious mechanisms that drive communication and reactions within human cells could be on the verge of being unravelled, due to a pioneering new technique.
Researchers from the Universities of Leeds, Exeter and Cambridge have harnessed an innovative new method to gain a greater understanding of signalling stations within the cells, called nanodomains.
They believe the new technique could pave the way for a greater understanding behind the causes of potentially life-threatening conditions such as heart disease, as well as potential new treatment pathways.
The nanodomains are known to drive fundamental physiological processes in the body, including the onset of disease.
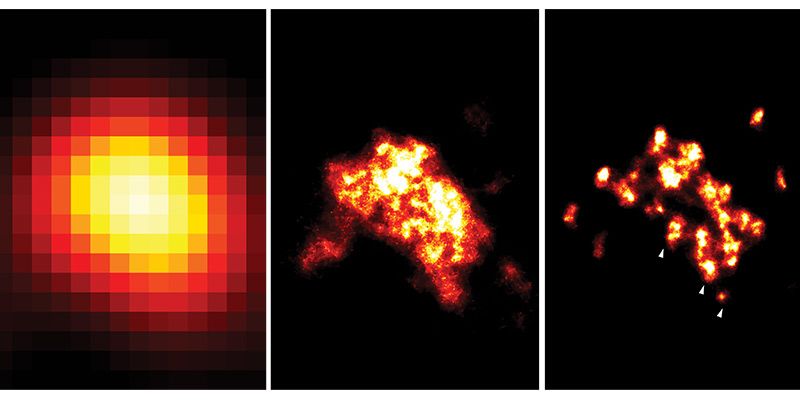
Improving technology: three images of a nanodomain visualised, from left to right, with TIRF (2006 technology), dSTORM (2009 technology) and DNA-PAINT (used in this study). The white arrowheads in the right image indicate individual proteins, which the team’s new technique allows them to identify. Image credit: Dr Isuru Jayasinghe
Scientists have, until now, generally relied on electron-microscopy to study these structures. However the technology has not allowed access to the finer mechanisms of the nanodomains at a molecular level.
Now, the UK research team have refined a new, light based super-resolution microscopy technique that allows high-quality imaging of the signalling stations in the human heart.
First author Dr Isuru Jayasinghe, from Leeds' School of Biomedical Sciences, said: “This new super-resolution microscopy tool gives us the perfect window to visually examine the individual protein changes within heart cells’ molecular machinery which lead to heart failure.
“At present, none of the treatments or therapies provided to heart failure patients specifically target the signalling stations – nanodomains – within the cell, which the evidence overwhelmingly suggests are a major cause of heart failure.
“We believe that by visualising these signalling structures at this level of detail using super-resolution microscopy we can help guide investigations into how we can target or repair these molecular machines and thus, in the long term, help patients to overcome heart disease.”
Expertise in the design of synthetic DNA strands was provided by Dr Lorenzo Di Michele from the University of Cambridge.
The ground-breaking new technique allows scientists to pin-point any number of specific types of proteins within the cells, the counting of each species of protein, and observations of the precise patterns in which they are arranged.
Professor Christian Soeller, who led the study and is at the new Living Systems Institute at the University of Exeter, said: “Slightly more than a decade ago nobody thought that we would ever see individual molecules with light, the resolution just seemed insufficient to resolve such fine detail.”
“Since then an astonishing array of new tricks has been devised. In our latest advance, the use of synthetic DNA has been critical – the deep understanding of the chemistry of DNA we have today makes it an enormously versatile tool.”
As a result, the team says that their research provides a “perfect window” to examine the changes that occur in the molecular machinery which are a major cause of heart failure.
They believe the added visual detail that the new imaging provides will guide more decisive investigations into how to target or repair these signalling stations or the molecular machines within them more precisely.
The study is published today in the journal Cell Reports.
Further information
For additional information and to request interviews with Dr Isuru Jayasinghe, please contact Simon Moore in the University of Leeds press office on +44 (0)113 34 34031 or s.i.moore@leeds.ac.uk.
The research paper True Molecular Scale Visualization of Variable Clustering Properties of Ryanodine Receptors is published 9 January 2018 in the journal Cell Reports.
The work was supported by funding from the Human Frontier Science Program, the Engineering and Physical Sciences Research Council of the UK, Wellcome Trust, and the Royal Society of the UK.