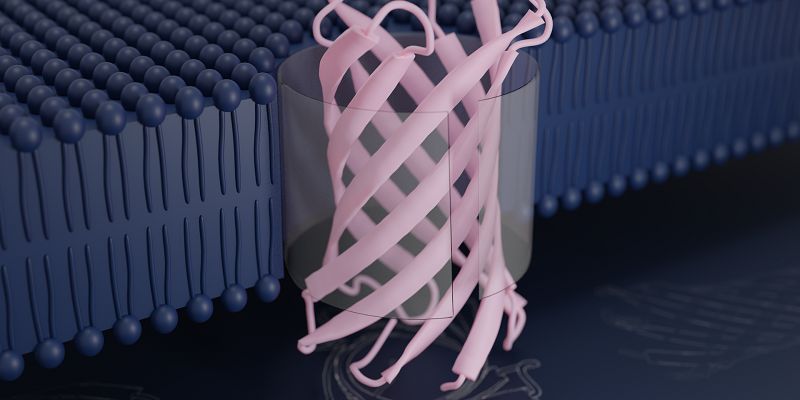
Scientists at Leeds are part of an international collaboration that has designed a protein that self-assembles into an artificial pore.
The protein sequence spontaneously transforms into a "transmembrane beta-barrel" – a tiny conduit or pore that embeds itself into a lipid membrane, mimicking its natural counterparts which are found in the walls of bacterial cells and enable nutrients, vitamins and strands of DNA to pass through.
There is intense scientific interest in the self-assembly of artificial pores as a way of exploiting the way nature moves "cargo" in and out of cells, for use in drug delivery or filtration technologies.
Professor Sheena Radford FRS is Director of the Astbury Centre for Structural Molecular Biology at Leeds and was co-supervisor of that part of the research conducted at the University.
She said: “Proteins are the molecular machines that carry out functions necessary to sustain life. Science has a lot to learn from the way they work, and there has been significant progress in designing bespoke proteins – but up to now, it has not been possible to design a protein that would form a transmembrane beta-barrel.
“What has been achieved in this study is a major advance.”
“These proteins are essential for bacterial growth, so if we can better understand how assemble, we might be better able to find new ways of beating bacterial infections...”
The study – De novo design of transmembrane β-barrels – has been published in the journal Science.
First author of the paper is Dr Anastassia Vorobieva, from the Institute for Protein Design led by Professor David Baker at the University of Washington School of Medicine. The Institute developed the Rosetta algorithm to design the 3-D structure of new proteins.
To create new transmembrane beta-barrels, Dr Vorobieva and colleagues used software to draft possible sequences that they predicted would fold to form the desired structures. They were building the molecule from scratch. Though they drew inspiration from proteins found in nature, they arrived at sequences that differ from any known before.
Their most successful designed proteins contain eight ribbon-like strands that fold into a compact barrel structure that stands just three nanometers tall, where a nanometre is one-billionth of a metre.
Dr Vorobieva said: “We began with a relatively simple notion about what would make the proteins fold. But when we tested these initial hypotheses, nothing worked at all. That was very frustrating.
“We didn’t assume we would get it right the first time, but we did think we could get some information back that would tell us how to move forward. Instead, I had to go back and look carefully at how nature solves this problem.
“The key was to try to detect patterns in those proteins. It was a really difficult thing to do.”
Leeds evaluated the protein design
The expertise at the Astbury Centre for Molecular and Structural Biology Leeds was brought in to validate that the new protein did in fact fold into a transmembrane beta barrel and embed itself into a lipid membrane. The analysis showed the protein could fold up efficiently without the help of any accessory proteins. This is in marked contrast to how natural transmembrane beta barrels fold.
David Brockwell, Professor of Biochemistry and Molecular Biology who also co-supervised the research at Leeds, said: “These designed proteins are interesting and immensely powerful from a basic science perspective because they have no evolutionary history. By studying them, we can discover some of the essential features that enable transmembrane beta-barrel proteins to fold into a membrane.
“Perhaps most importantly, these type of proteins are essential for bacterial growth and survival, so if we can better understand how they fold and assemble, we might be better able to find new ways of beating bacterial infections with new antibiotics.”
This work is the latest achievement in the rapidly progressing field of protein design. In recent years, scientists at the Institute for Protein Design have created innovative vaccines, experimental cancer treatments, and sensors capable of detecting antibodies against COVID-19.
Further information
Image courtesy: Institute for Protein Design
The research was supported by the Medical Research Council, Wellcome Trust, Biotechnology and Biological Sciences Research Council, National Institutes of Health, Howard Hughes Medical Institute, Fulbright Belgium and Luxembourg, Open Philanthropy Project, Air Force Office of Scientific Research, Nordstrom Barrier Fund, and Eric and Wendy Schmidt by recommendation of the Schmidt Futures program.
For further details, please email David Lewis in the university press office at: d.lewis@leeds.ac.uk